Question 1: Find out the sequence weights for the sequences ACTA, ACTT, CGTT and AGAT by using Thompson, Higgins and Gibson method
a) Compute pair wise distances between the sequences.
b) Build phylogenetic the tree by using UPGMA method.
c) Derive sequence weights.
Question 2: We supposed additive property when constructed UPGMA tree in question 1. What is limitation of this supposition?
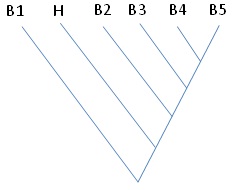
Question 3: The sequence of gene ‘B3’ was used to blast against swissprot protein database. The query returned five significant hits to the other proteins denoted by B1, B2, B4, B5 and H in the figure shown below. B1-B5 is five bacterial species, while the species H represents Homo sapiens. No other mammalian species have shown presence of protein that is similar to B3. Describe the presence of this gene in the humans.
Problem 4: Explain technical and theoretical challenges related with building phylogenetic trees.