Determine sequence weights for the sequences ACTA, ACTT, CGTT, and AGAT in problem 1 by using Thompson, Higgins, and Gibson method
a) compute pairwise distances between sequences
b) build phylogenetic the tree using UPGMA method
c) derive sequence weights
Problem 2
We assumed additive property when constructed UPGMA tree in problem 1.
What is limitation of this assumption?
Problem 3
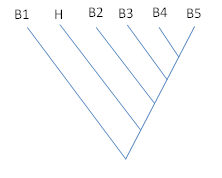
The sequence of gene "B3" was used to blast against swissprot protein database. The query returned five significant hits to other proteins denoted by B1,B2,B4, B5, and H in the figure below. B1-B5 are five bacterial species, while the species H represents Homo sapiens. No other mammalian species have shown presence of protein that is similar to B3. Explain the presence of this gene in humans.
Problem 4
Describe technical and theoretical challenges associated with building phylogenetic trees.